Example: One Max¶
We will consider the OneMax problem as an example of a simple problem solvable with Genetic Algorithms. Given a sequence N of random bits composing a word (i.e. 10001010 8 bits), the fitness score is given based on the number of ones present (higher is better). The algorithm must generate a random set of individuals (strings of bits), evolve them till they will contain only ones.
Base¶
In version one an individual is composed by a word_section which contains
a single macro (word_macro) with a parameter (bit) of type
microgp.parameter.bitstring.Bitstring of length 8 bits. The main
section contains a simple prologue and epilogue.
The evaluator in both versions is a Python method (evaluator_function) returning an int value
that is the sum of 1 in the individual’s phenotype.
import argparse
import sys
import microgp as ugp
from microgp.utils import logging
if __name__ == "__main__":
ugp.banner()
parser = argparse.ArgumentParser()
parser.add_argument("-v", "--verbose", action="count", default=0, help="increase log verbosity")
parser.add_argument("-d", "--debug", action="store_const", dest="verbose", const=2,
help="log debug messages (same as -vv)")
args = parser.parse_args()
if args.verbose == 0:
ugp.logging.DefaultLogger.setLevel(level=ugp.logging.INFO)
elif args.verbose == 1:
ugp.logging.DefaultLogger.setLevel(level=ugp.logging.VERBOSE)
elif args.verbose > 1:
ugp.logging.DefaultLogger.setLevel(level=ugp.logging.DEBUG)
ugp.logging.debug("Verbose level set to DEBUG")
ugp.logging.cpu_info("Program started")
# Define a parameter of type ugp.parameter.Bitstring and length = 8
word8 = ugp.make_parameter(ugp.parameter.Bitstring, len_=8)
# Define a macro that contains a parameter of type ugp.parameter.Bitstring
word_macro = ugp.Macro("{word8}", {'word8': word8})
# Create a section containing a macro
word_section = ugp.make_section(word_macro, size=(1, 1), name='word_sec')
# Create a constraints library
library = ugp.Constraints()
# Define the sections in the library
library['main'] = ["Bitstring:", word_section]
# Define the evaluator method and the fitness type
def evaluator_function(data: str):
count = data.count('1')
return list(str(count))
library.evaluator = ugp.fitness.make_evaluator(evaluator=evaluator_function, fitness_type=ugp.fitness.Lexicographic)
# Create a list of operators with their aritiy
operators = ugp.Operators()
# Add initialization operators
operators += ugp.GenOperator(ugp.create_random_individual, 0)
# Add mutation operators
operators += ugp.GenOperator(ugp.hierarchical_mutation, 1)
operators += ugp.GenOperator(ugp.flat_mutation, 1)
# Create the object that will manage the evolution
mu = 10
nu = 20
sigma = 0.7
lambda_ = 7
max_age = 10
darwin = ugp.Darwin(
constraints=library,
operators=operators,
mu=mu,
nu=nu,
lambda_=lambda_,
sigma=sigma,
max_age=max_age,
)
# Evolve and print individuals in population
darwin.evolve()
logging.bare("This is the final population:")
for individual in darwin.population:
msg = f"Solution {str(individual.node_id)} "
ugp.print_individual(individual, msg=msg, plot=True)
ugp.logging.bare(f"Fitness: {individual.fitness}")
ugp.logging.bare("")
# Print best individuals
ugp.print_individual(darwin.archive.individuals, msg="These are the best ever individuals:", plot=True)
ugp.logging.cpu_info("Program completed")
sys.exit(0)
Structured¶
In version two an individual is composed by a word_section which contains
exactly 8 macros (word_macro) with a parameter (bit) of type
microgp.parameter.categorical.Categorical that can assume as value: 1 or
0. The main section contains a simple prologue and epilogue.
import argparse
import sys
import microgp as ugp
from microgp.utils import logging
if __name__ == "__main__":
ugp.banner()
parser = argparse.ArgumentParser()
parser.add_argument("-v", "--verbose", action="count", default=0, help="increase log verbosity")
parser.add_argument("-d", "--debug", action="store_const", dest="verbose", const=2,
help="log debug messages (same as -vv)")
args = parser.parse_args()
if args.verbose == 0:
ugp.logging.DefaultLogger.setLevel(level=ugp.logging.INFO)
elif args.verbose == 1:
ugp.logging.DefaultLogger.setLevel(level=ugp.logging.VERBOSE)
elif args.verbose > 1:
ugp.logging.DefaultLogger.setLevel(level=ugp.logging.DEBUG)
ugp.logging.debug("Verbose level set to DEBUG")
ugp.logging.cpu_info("Program started")
# Define a parameter of type ugp.parameter.Categorical that can take two values: 0 or 1
bit = ugp.make_parameter(ugp.parameter.Categorical, alternatives=[0, 1])
# Define a macro that contains a parameter of type ugp.parameter.Categorical
word_macro = ugp.Macro("{bit}", {'bit': bit})
# Create a section containing 8 macros
word_section = ugp.make_section(word_macro, size=(8, 8), name='word_sec')
# Create a constraints library
library = ugp.Constraints()
library['main'] = ["Bitstring:", word_section]
# Define the evaluator method and the fitness type
def evaluator_function(data: str):
count = data.count('1')
return list(str(count))
library.evaluator = ugp.fitness.make_evaluator(evaluator=evaluator_function, fitness_type=ugp.fitness.Lexicographic)
# Create a list of operators with their arity
operators = ugp.Operators()
# Add initialization operators
operators += ugp.GenOperator(ugp.create_random_individual, 0)
# Add mutation operators
operators += ugp.GenOperator(ugp.hierarchical_mutation, 1)
operators += ugp.GenOperator(ugp.flat_mutation, 1)
# Add crossover operators
operators += ugp.GenOperator(ugp.macro_pool_one_cut_point_crossover, 2)
operators += ugp.GenOperator(ugp.macro_pool_uniform_crossover, 2)
# Create the object that will manage the evolution
mu = 10
nu = 20
sigma = 0.7
lambda_ = 7
max_age = 10
darwin = ugp.Darwin(
constraints=library,
operators=operators,
mu=mu,
nu=nu,
lambda_=lambda_,
sigma=sigma,
max_age=max_age,
)
# Evolve and print individuals in population
darwin.evolve()
logging.bare("This is the final population:")
for individual in darwin.population:
msg = f"Solution {str(individual.node_id)} "
ugp.print_individual(individual, msg=msg, plot=True, score=True)
# Print best individuals
ugp.print_individual(darwin.archive.individuals, msg="These are the best ever individuals:", plot=True)
ugp.logging.cpu_info("Program completed")
sys.exit(0)
Assembly¶
The following code produces assembly code that can be run on x86 processors.
The goal is to generate an assembly script that writes in eax a binary
number with as much as ones (1) as possible.
The evaluator is a .bat file that generates an .exe file in charge
of call the script and count the number of ones in the returned integer
value.
@echo off
rem comment
del a.exe
gcc main.o %1
if exist a.exe (
.\a.exe
) else (
echo -1
)
A possible solution could be:
.file "solution.c"
.text
.globl _darwin
.def _darwin; .scl 2; .type 32; .endef
_darwin:
LFB17:
.cfi_startproc
pushl %ebp
.cfi_def_cfa_offset 8
.cfi_offset 5, -8
movl %esp, %ebp
.cfi_def_cfa_register 5
movl $-31312, %eax
movl $25598, %ebx
movl $-24861, %ecx
movl $-19236, %edx
sub %ebx, %edx
shl $216, %ecx
jnz n9
jnc n23
cmp %ecx, %ecx
shl $207, %edx
n9:
jc n22
xor %ebx, %eax
jnz n28
xor %eax, %ebx
sub %edx, %edx
jno n15
n15:
jz n28
shr $229, %ebx
sub %ebx, %eax
jc n23
cmp %edx, %ebx
and %ebx, %ecx
shl $186, %eax
n22:
cmp %eax, %edx
n23:
jnz n29
jz n29
jmp n28
jc n29
shl $143, %ecx
n28:
or %ebx, %eax
n29:
movl %eax, -4(%ebp)
movl -4(%ebp), %eax
leave
.cfi_restore 5
.cfi_def_cfa 4, 4
ret
.cfi_endproc
LFE17:
.ident "GCC: (MinGW.org GCC-8.2.0-5) 8.2.0"
Fitness score: Lexicographic(29)
The correspondent graph_manager plot is:
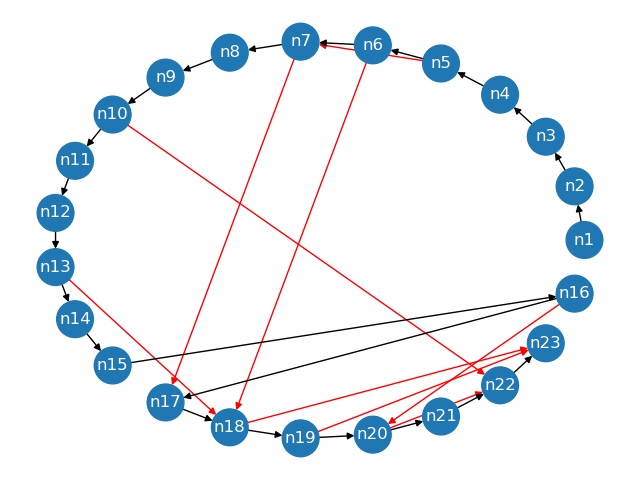
In the figure the black edges are next edges and the red ones are LocalReferences (jump).
import argparse
import sys
import microgp as ugp
from microgp.utils import logging
if __name__ == "__main__":
ugp.banner()
parser = argparse.ArgumentParser()
parser.add_argument("-v", "--verbose", action="count", default=0, help="increase log verbosity")
parser.add_argument("-d", "--debug", action="store_const", dest="verbose", const=2,
help="log debug messages (same as -vv)")
args = parser.parse_args()
if args.verbose == 0:
ugp.logging.DefaultLogger.setLevel(level=ugp.logging.INFO)
elif args.verbose == 1:
ugp.logging.DefaultLogger.setLevel(level=ugp.logging.VERBOSE)
elif args.verbose > 1:
ugp.logging.DefaultLogger.setLevel(level=ugp.logging.DEBUG)
ugp.logging.debug("Verbose level set to DEBUG")
ugp.logging.cpu_info("Program started")
# Define parameters
reg_alternatives = ['%eax', '%ebx', '%ecx', '%edx']
reg_param = ugp.make_parameter(ugp.parameter.Categorical, alternatives=reg_alternatives)
instr_alternatives = ['add', 'sub', 'and', 'or', 'xor', 'cmp']
instr_param = ugp.make_parameter(ugp.parameter.Categorical, alternatives=instr_alternatives)
shift_alternatives = ['shr', 'shl']
shift_param = ugp.make_parameter(ugp.parameter.Categorical, alternatives=shift_alternatives)
jmp_alternatives = ['ja', 'jz', 'jnz', 'je', 'jne', 'jc', 'jnc', 'jo', 'jno', 'jmp']
jmp_instructions = ugp.make_parameter(ugp.parameter.Categorical, alternatives=jmp_alternatives)
integer = ugp.make_parameter(ugp.parameter.Integer, min=-32768, max=32767)
int8 = ugp.make_parameter(ugp.parameter.Integer, min=0, max=256)
jmp_target = ugp.make_parameter(ugp.parameter.LocalReference,
allow_self=False,
allow_forward=True,
allow_backward=False,
frames_up=0)
# Define the macros
jmp1 = ugp.Macro(" {jmp_instr} {jmp_ref}", {'jmp_instr': jmp_instructions, 'jmp_ref': jmp_target})
instr_op_macro = ugp.Macro(" {instr} {regS}, {regD}",{'instr': instr_param, 'regS': reg_param, 'regD': reg_param})
shift_op_macro = ugp.Macro(" {shift} ${int8}, {regD}", {'shift': shift_param, 'int8': int8, 'regD': reg_param})
branch_macro = ugp.Macro("{branch} {jmp}", {'branch': jmp_instructions, 'jmp': jmp_target})
prologue_macro = ugp.Macro(' .file "solution.c"\n' +
' .text\n' +
' .globl _darwin\n' +
' .def _darwin; .scl 2; .type 32; .endef\n' +
'_darwin:\n' +
'LFB17:\n' +
' .cfi_startproc\n' +
' pushl %ebp\n' +
' .cfi_def_cfa_offset 8\n' +
' .cfi_offset 5, -8\n' +
' movl %esp, %ebp\n' +
' .cfi_def_cfa_register 5\n')
init_macro = ugp.Macro(" movl ${int_a}, %eax\n" +
" movl ${int_b}, %ebx\n" +
" movl ${int_c}, %ecx\n" +
" movl ${int_d}, %edx\n",
{'int_a': integer, 'int_b': integer, 'int_c': integer, 'int_d': integer})
epilogue_macro = ugp.Macro(
' movl %eax, -4(%ebp)\n' +
' movl -4(%ebp), %eax\n' +
' leave\n' +
' .cfi_restore 5\n' +
' .cfi_def_cfa 4, 4\n' +
' ret\n' +
' .cfi_endproc\n' +
'LFE17:\n' +
' .ident "GCC: (MinGW.org GCC-8.2.0-5) 8.2.0"\n')
# Define section
sec1 = ugp.make_section({jmp1, instr_op_macro, shift_op_macro}, size=(1, 50))
# Create a constraints library
library = ugp.Constraints(file_name="solution{node_id}.s")
library['main'] = [prologue_macro, init_macro, sec1, epilogue_macro]
# Define the evaluator script and the fitness type
if sys.platform != "win32":
exit(-1)
else:
script = "eval.bat"
library.evaluator = ugp.fitness.make_evaluator(evaluator=script, fitness_type=ugp.fitness.Lexicographic)
# Create a list of operators with their arity
operators = ugp.Operators()
# Add initialization operators
operators += ugp.GenOperator(ugp.create_random_individual, 0)
# Add mutation operators
operators += ugp.GenOperator(ugp.hierarchical_mutation, 1)
operators += ugp.GenOperator(ugp.flat_mutation, 1)
operators += ugp.GenOperator(ugp.add_node_mutation, 1)
operators += ugp.GenOperator(ugp.remove_node_mutation, 1)
# Add crossover operators
operators += ugp.GenOperator(ugp.macro_pool_one_cut_point_crossover, 2)
operators += ugp.GenOperator(ugp.macro_pool_uniform_crossover, 2)
# Create the object that will manage the evolution
mu = 10
nu = 20
sigma = 0.7
lambda_ = 7
max_age = 10
darwin = ugp.Darwin(
constraints=library,
operators=operators,
mu=mu,
nu=nu,
lambda_=lambda_,
sigma=sigma,
max_age=max_age,
)
# Evolve
darwin.evolve()
# Print best individuals
logging.bare("These are the best ever individuals:")
best_individuals = darwin.archive.individuals
ugp.print_individual(best_individuals, plot=True, score=True)
ugp.logging.cpu_info("Program completed")
sys.exit(0)
The script syntax has been built to work with Windows 10, 64-bit, for GCC: (MinGW.org GCC-8.2.0-5) 8.2.0.